Note
Go to the end to download the full example code.
Matrix Contribution Correlation Comparison
This example demonstrates how to generate a matrix comparison plot for the contribution correlation method (described in Matrix of Contribution Correlation Plots and Perturbation Correlation Plots). This example compares the calculated correlation correlation coefficient to the TSUNAMI-IP calculated \(c_k\) value for a set of SDFs computed from the HMF series of critical experiments [Bess2019].
Getting TSUNAMI-IP \(c_k\) Values
First, we need to get the TSUNAMI-IP \(c_k\) values for the experiment and application SDFs we want to compare. We can do this
by using the tsunami_ip_utils.integral_indices.get_integral_indices() function, as shown in
Reading Integral Indices.
from tsunami_ip_utils.integral_indices import get_integral_indices
from paths import EXAMPLES
application_sdfs = [ EXAMPLES / 'data' / 'example_sdfs' / 'HMF' / f'HEU-MET-FAST-003-00{i}.sdf' for i in range(1, 8) ]
experiment_sdfs = application_sdfs
# Get the TSUNAMI-IP integral indices
coverx_library = '56groupcov7.1'
integral_indices = get_integral_indices(application_sdfs, experiment_sdfs, coverx_library=coverx_library)
c_k = integral_indices['c_k']
Nuclide-Wise Comparison
Variance Contributions
First we show a comparison of the calculated correlation coefficient using the contribution correlation method with nuclide-wise
variance contributions to the TSUNAMI-IP \(c_k\) values. To generate this comparison, we just have to specify the correct method,
which in this case is 'variance_contributions_nuclide'.
from tsunami_ip_utils.comparisons import correlation_comparison
labels = {
'applications': [ application_file.name for application_file in application_sdfs ],
'experiments': [ experiment_file.name for experiment_file in experiment_sdfs ],
}
comparisons, fig = correlation_comparison(
integral_index_matrix=c_k,
integral_index_name='c_k',
application_files=application_sdfs,
experiment_files=experiment_sdfs,
method='variance_contributions_nuclide',
matrix_plot_kwargs={'labels': labels},
)
fig.show()
Uncertainty Contributions
We can also generate a comparison using the contribution correlation method with the uncertainty contributions
comparisons, fig = correlation_comparison(
integral_index_matrix=c_k,
integral_index_name='c_k',
application_files=application_sdfs,
experiment_files=experiment_sdfs,
method='uncertainty_contributions_nuclide',
)
fig.show()
We can also generate an image of the comparison
comparisons, fig = correlation_comparison(
integral_index_matrix=c_k,
integral_index_name='c_k',
application_files=application_sdfs,
experiment_files=experiment_sdfs,
method='uncertainty_contributions_nuclide',
)
fig.to_image( EXAMPLES / '_static' / 'contribution_matrix_comparison.png' )
# sphinx_gallery_thumbnail_path = '../../examples/_static/contribution_matrix_comparison.png'
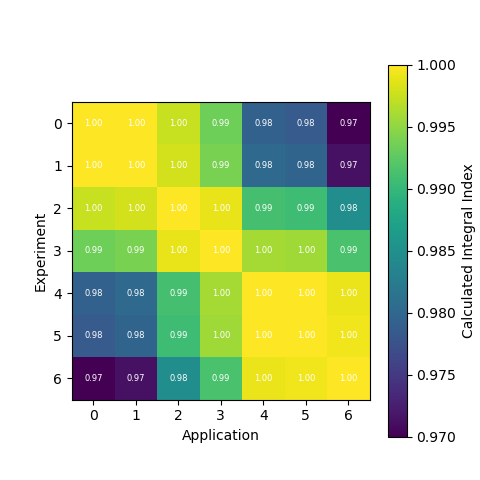
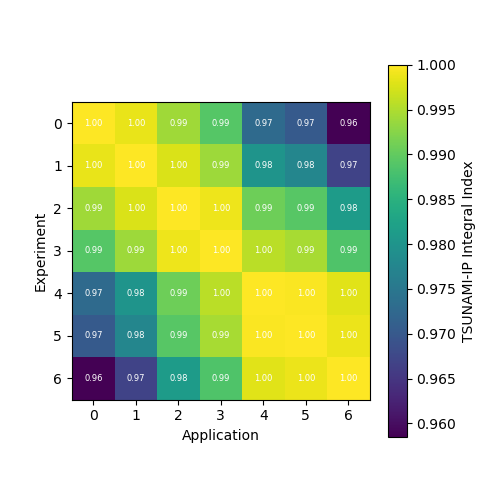
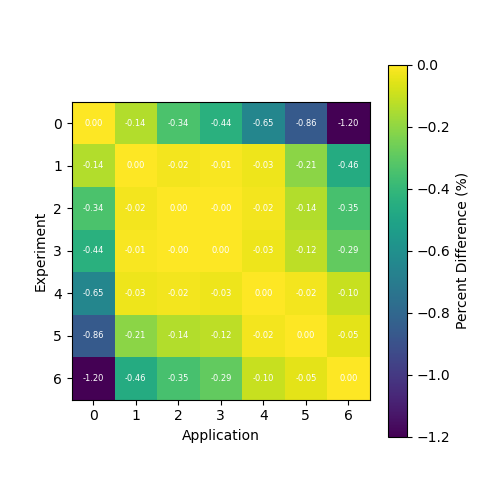
Generating a Comparison Heatmap
We can also generate a heatmap of the comparison between the calculated correlation coefficient and the TSUNAMI-IP \(c_k\) values
by using the tsunami_ip_utils.viz.generate_heatmap_from_comparison() function.
from tsunami_ip_utils.viz import generate_heatmap_from_comparison
plot_dict = generate_heatmap_from_comparison(comparisons)
print(plot_dict.keys())
dict_keys(['Calculated', 'TSUNAMI-IP', 'Percent Difference'])
The tsunami_ip_utils.viz.generate_heatmap_from_comparison() takes in the comparison dataframe generated by the
tsunami_ip_utils.comparisons_correlation_comparison() function and returns a dictionary of matplotlib heatmaps. From the
output above you can see that the function contains heatmaps of the calculated value, the TSUNAMI-IP value, and the percent
difference between the two. Note that this function was used to generate the plots shown in the technical manual.
Total running time of the script: (2 minutes 4.479 seconds)