Note
Go to the end to download the full example code.
SCALE Multigroup Cross Section Reader
This example demonstrates how to read SCALE multigroup cross section libraries. This functionality relies on AMPX modules for creating text dumps of COVERX formatted libraries. More details are given here: Multigroup SCALE XS Library Reader.
A Disclaimer
The COVERX formatted cross section library referenced in this example dummy_56_v7.1 was created with AMPX using ExSite
and publicly available cross section data from ENDF VII.1.
This library is NOT intended for use in simulations, and only contains a small subset of nuclides and reactions. This
library is included purely for demonstration purposes, and when using functionality in this package that requires nuclear
data, please use the provided SCALE libraries.
Reading Multigroup Cross Sections
The following code snippet demonstrates how to read a SCALE multigroup cross section library. The function
tsunami_ip_utils.xs.read_multigroup_xs() is used to read COVER formatted SCALE cross section libraries, and takes
a pathlib.Path object to the multigroup library, and a dictionary of nuclide-reaction pairs to read. The function
returns a nested dictionary of numpy.ndarray objects, where the outer dictionary keys are nuclides, and the inner
dictionary keys are reactions. The values are the multigroup cross sections for the corresponding nuclide-reaction pair.
from tsunami_ip_utils.xs import read_multigroup_xs
from paths import EXAMPLES
nuclide_reaction_dict = {'92235': ['1', '18'], '5011': ['1', '27'], '94239': ['1', '18']}
multigroup_library_path = EXAMPLES / 'data' / 'dummy_56_v7.1'
out = read_multigroup_xs(multigroup_library_path, nuclide_reaction_dict)
print(len(out['92235']['1']))
56
This function is parallel, and reads cross section libraries on multiple cores, which can be useful for large libraries.
This function isn’t the most user-friendly, and requires the user to input a nuclide_reaction_dict in terms of nuclide
ZAID and reaction MT numbers, but it is primarily used by the tsunami_ip_utils.perturbations module for
reading cross sections for perturbation calculations.
Caching a Multigroup Cross Section Library
If working with python-based applications that deal with SCALE multigroup cross section data, it may be useful to cache the
cross section libary in a convenient format for reading into python, like a .pkl file. To avoid having to manually supply
the list of all nuclides and reactions in the multigroup library, the tsunami_ip_utils.xs.read_multigroup_xs() function
can be run with the return_available_nuclide_reactions flag set to True. This will return a dictionary of all nuclides
and reactions in the library, which can then be used to read the library and cache it.
out, available_nuclide_reactions = read_multigroup_xs(
multigroup_library_path,
nuclide_reaction_dict,
return_available_nuclide_reactions=True
)
print(available_nuclide_reactions)
{'5011': ['1099', '3099', '2', '4', '16', '22', '28', '51', '52', '53', '54', '55', '56', '57', '58', '59', '60', '91', '102', '103', '105', '107', '1', '3002', '3102', '101', '27', '3', '1007'], '92235': ['1099', '3099', '2', '3', '4', '16', '17', '18', '1452', '452', '1456', '456', '1455', '455', '19', '1419', '4561', '20', '1420', '4562', '21', '1421', '4563', '37', '38', '1438', '4564', '51', '52', '53', '54', '55', '56', '57', '58', '59', '60', '61', '62', '63', '64', '65', '66', '67', '68', '69', '70', '71', '72', '73', '74', '75', '76', '77', '78', '79', '80', '81', '82', '83', '84', '85', '86', '87', '88', '89', '90', '91', '102', '1', '3002', '3018', '3102', '101', '27', '1018', '1056', '1055', '1019', '1020', '1021', '1038', '1007'], '94239': ['1099', '3099', '2', '3', '4', '16', '17', '18', '1452', '452', '1456', '456', '1455', '455', '19', '1419', '4561', '20', '1420', '4562', '21', '1421', '4563', '37', '38', '1438', '4564', '51', '52', '53', '54', '55', '56', '57', '58', '59', '60', '61', '62', '63', '64', '65', '66', '67', '68', '69', '70', '71', '72', '73', '74', '75', '76', '77', '78', '79', '80', '81', '82', '83', '84', '85', '86', '87', '88', '89', '90', '91', '102', '1', '3002', '3018', '3102', '101', '27', '1018', '1056', '1055', '1019', '1020', '1021', '1038', '1007']}
The list of all available nuclide reactions can then be supplied to the function to read the entire library.
out = read_multigroup_xs(multigroup_library_path, available_nuclide_reactions)
And the cross sections can be easily cached
import pickle
with open(EXAMPLES / 'data' / 'dummy_56_v7.1.pkl', 'wb') as f:
pickle.dump(out, f)
Getting the Energy Structure
By default the reader doesn’t return the energy group structure (this is something that could be improved in the future), but if
you’re examining SCALE built-in cross section libraries, the energy group structure can be obtained from the
tsunami_ip_utils.xs.get_scale_multigroup_structure() function. This function scrapes the SCALE manual for the energy
group structure of a given SCALE library.
from tsunami_ip_utils.xs import get_scale_multigroup_structure
num_groups = 56
multigroup_structure = get_scale_multigroup_structure(num_groups)
print(multigroup_structure)
[[5.600e+01 4.000e-03]
[5.500e+01 1.000e-02]
[5.400e+01 2.530e-02]
[5.300e+01 4.000e-02]
[5.200e+01 5.000e-02]
[5.100e+01 6.000e-02]
[5.000e+01 8.000e-02]
[4.900e+01 1.000e-01]
[4.800e+01 1.500e-01]
[4.700e+01 2.000e-01]
[4.600e+01 2.500e-01]
[4.500e+01 3.250e-01]
[4.400e+01 3.500e-01]
[4.300e+01 3.750e-01]
[4.200e+01 4.500e-01]
[4.100e+01 6.250e-01]
[4.000e+01 1.010e+00]
[3.900e+01 1.080e+00]
[3.800e+01 1.130e+00]
[3.700e+01 5.000e+00]
[3.600e+01 6.250e+00]
[3.500e+01 6.500e+00]
[3.400e+01 6.875e+00]
[3.300e+01 7.000e+00]
[3.200e+01 2.050e+01]
[3.100e+01 2.120e+01]
[3.000e+01 2.175e+01]
[2.900e+01 3.600e+01]
[2.800e+01 3.713e+01]
[2.700e+01 6.500e+01]
[2.600e+01 6.750e+01]
[2.500e+01 1.012e+02]
[2.400e+01 1.050e+02]
[2.300e+01 1.160e+02]
[2.200e+01 1.175e+02]
[2.100e+01 1.877e+02]
[2.000e+01 1.915e+02]
[1.900e+01 2.250e+03]
[1.800e+01 3.740e+03]
[1.700e+01 1.700e+04]
[1.600e+01 2.000e+04]
[1.500e+01 5.000e+04]
[1.400e+01 2.000e+05]
[1.300e+01 2.700e+05]
[1.200e+01 3.300e+05]
[1.100e+01 4.700e+05]
[1.000e+01 6.000e+05]
[9.000e+00 7.500e+05]
[8.000e+00 8.611e+05]
[7.000e+00 1.200e+06]
[6.000e+00 1.500e+06]
[5.000e+00 1.850e+06]
[4.000e+00 3.000e+06]
[3.000e+00 4.304e+06]
[2.000e+00 6.434e+06]
[1.000e+00 2.000e+07]]
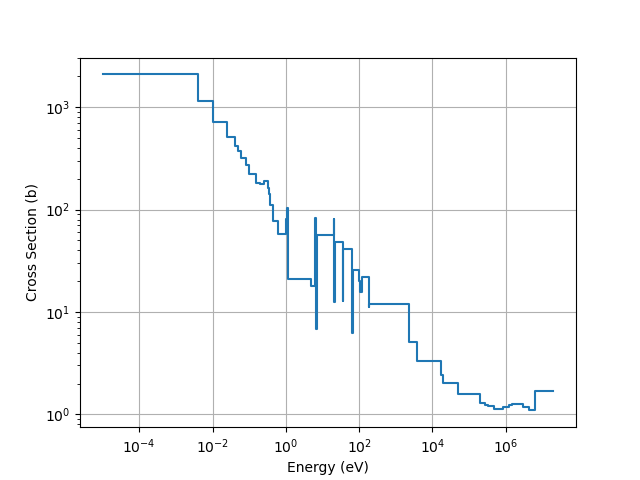
We can easily plot cross section data using the energy group structure. The following code snippet demonstrates how to plot the u-235 fission cross section
import matplotlib.pyplot as plt
import numpy as np
u235_fission_xs = out['92235']['18']
# To get the cross sections to be visually constant within each energy group, we need to repeat the cross sections and add in the
# implied lower energy bound of 1E-05 eV
u235_fission_xs = u235_fission_xs.repeat(2)
modified_energies = np.zeros_like(u235_fission_xs)
energies = multigroup_structure[:, 1]
modified_energies[0] = 1E-05
modified_energies[-1] = energies[-1]
modified_energies[1:] = np.repeat(energies, 2)[:-1]
plt.plot(modified_energies, u235_fission_xs)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Energy (eV)')
plt.ylabel('Cross Section (b)')
plt.grid()
plt.show()
A Future Improvement
It is unfortunate that to cache a cross section library, the library must be read twice (the most consuming part is making
the text dump again). This is a limitation of the current implementation, and may be improved in the future. To avoid this,
the text dump just needs to be saved so that it can be read by the second function call. This could all be implemented by
adding an additional flag to the tsunami_ip_utils.xs.read_multigroup_xs() that does this under the hood.
Total running time of the script: (0 minutes 9.919 seconds)